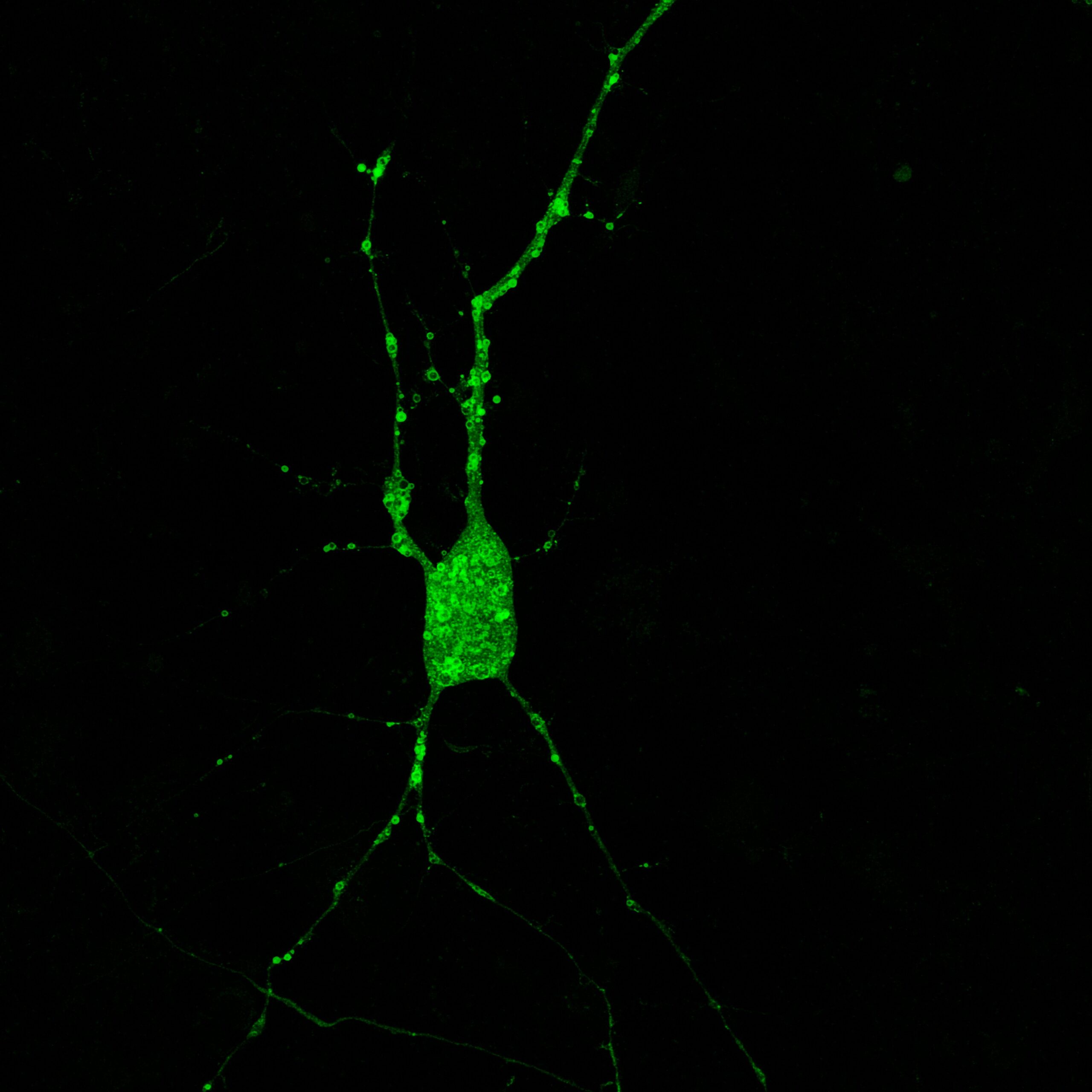
Visualize proteostasis and autophagy processes in living cells with a fluorescent probe
Green OmegaPaint is a new addition to our suite of Cell Painting tools, combining the bright mNeonGreen fluorescent protein with a portion of the DFCP1 protein (Double FYVE Containing Protein One) (Gillooly et al., 2001). DFCP1 defines the omegasome, and contains two domains that specifically interact with the phosphatidylinositol 3-phosphate (PIP3) that is produced by the PI3-kinase VPS34 (Nähse et al., 2024). This unique kinase regulates autophagy, and without its activity phagosome formation is blocked. The omegasome membranes and structures are abundant and quite mobile in living cells, and the omegasome paint makes it possible to watch proteostasis in real time. Please note that these are small structures that are best resolved using high numerical aperture objectives and confocal optics that reject out of focus light.
Autophagy is a fundamental cellular process that is crucial for maintaining cellular homeostasis. During autophagy, double membrane vesicles engulf and remove cellular debris, such as dead organelles and protein aggregates. In turn, it generates lipids, sugars, and amino acids for cellular life. A fascinating question in the field of autophagy has long involved the source of the membranes that make up the autophagosome. Evidence has been marshalled for many possible sources including the ERGIC compartment, the endoplasmic reticulum, the mitochondria, and the plasma membrane (Nähse et al., 2024). The most compelling evidence to date involves the endoplasmic reticulum, though it is not necessarily the sole source.
Figure 1: Green OmegaPaint used to visualize autophagasomes in an iPSC-derived, cortical glutamatergic neuron from BrainXell.
While many molecular details are only now emerging on the formation of autophagosomes, there are some key details that are well established. First, the PI3K kinase VSP34 is clearly a regulator of the process. This enzyme produces PI3P in the endoplasmid reticulum, where it is not normally found. Patches of this lipid recruit a number of omegasome relevant proteins, most notably the ATPase DFCP1 (Nähse et al., 2023). The mTOR complex has long been known to regulate autophagy, and it appears that it directly controls the PI3K kinase VSP34 activity (Yuan et al., 2013). As patches of the PI3P recruit key proteins bulges appear to be omega shaped, which is why these very early phagosomes are now referred to as the “omegasome.” Much remains to be learned about the omegasome in terms of its regulation, trafficking and molecular components.
References
Axe, E. L., Walker, S. A., Manifava, M., Chandra, P., Roderick, H. L., Habermann, A., Griffiths, G., & Ktistakis, N. T. (2008). Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. The Journal of Cell Biology, 182(4), 685–701. https://doi.org/10.1083/jcb.200803137
Gillooly, D. J., Simonsen, A., & Stenmark, H. (2001). Cellular functions of phosphatidylinositol 3-phosphate and FYVE domain proteins. Biochemical Journal, 355(Pt 2), 249–258. https://doi.org/10.1042/0264-6021:3550249
Nähse, V., Raiborg, C., Tan, K. W., Mørk, S., Torgersen, M. L., Wenzel, E. M., Nager, M., Salo, V. T., Johansen, T., Ikonen, E., Schink, K. O., & Stenmark, H. (2023). ATPase activity of DFCP1 controls selective autophagy. Nature Communications, 14(1), 4051. https://doi.org/10.1038/s41467-023-39641-9
Nähse, V., Stenmark, H., & Schink, K. O. (2024). Omegasomes control formation, expansion, and closure of autophagosomes. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology, 46(6), e2400038. https://doi.org/10.1002/bies.202400038
Yuan, H.-X., Russell, R. C., & Guan, K.-L. (2013). Regulation of PIK3C3/VPS34 complexes by MTOR in nutrient stress-induced autophagy. Autophagy, 9(12), 1983–1995. https://doi.org/10.4161/auto.26058
BacMam Gene Delivery
Custom BacMam preparations to deliver your gene of interest to mammalian cells.