Next-generation drugs for the evolving SARS-CoV-2 virus
There is increasing evidence that new variants of SARS-CoV-2 are appearing that cannot be treated with Paxlovid. This means that the next generation of antivirals will soon be needed to reduce the severity of COVID-19 infection. Recently, Pérez-Vargas and colleagues published a comprehensive, multidisciplinary study that reports promising new inhibitors to address the need for next-generation antivirals (Pérez-Vargas et al., 2023).
Replication of the SARS-CoV-2 virus depends upon its main protease, 3CLpro. 3CLpro works by cleaving the viral polyprotein into the many small parts necessary for virus replication. Because this protease is essential and conserved between many strains of coronavirus, it is an important drug target (Smith et al., 2023). If an inhibitor can block the protease, viral replication can be stopped. Paxlovid contains Nirmatrelvir, an inhibitor of the SARS-CoV-2 3CLpro (Joyce et al., 2022). This drug has been successful in treating infections, and is an important antiviral for treating at-risk patients (Owen et al., 2021). Unfortunately, only a few mutations in the 3CLpro are sufficient to render Nirmatrelvir ineffective (Iketani et al., 2023; Kiso et al., 2023; Moghadasi et al., 2023). These mutations are present now in rare strains of the virus, but it’s only a matter of time and evolution until a new inhibitor will be necessary.
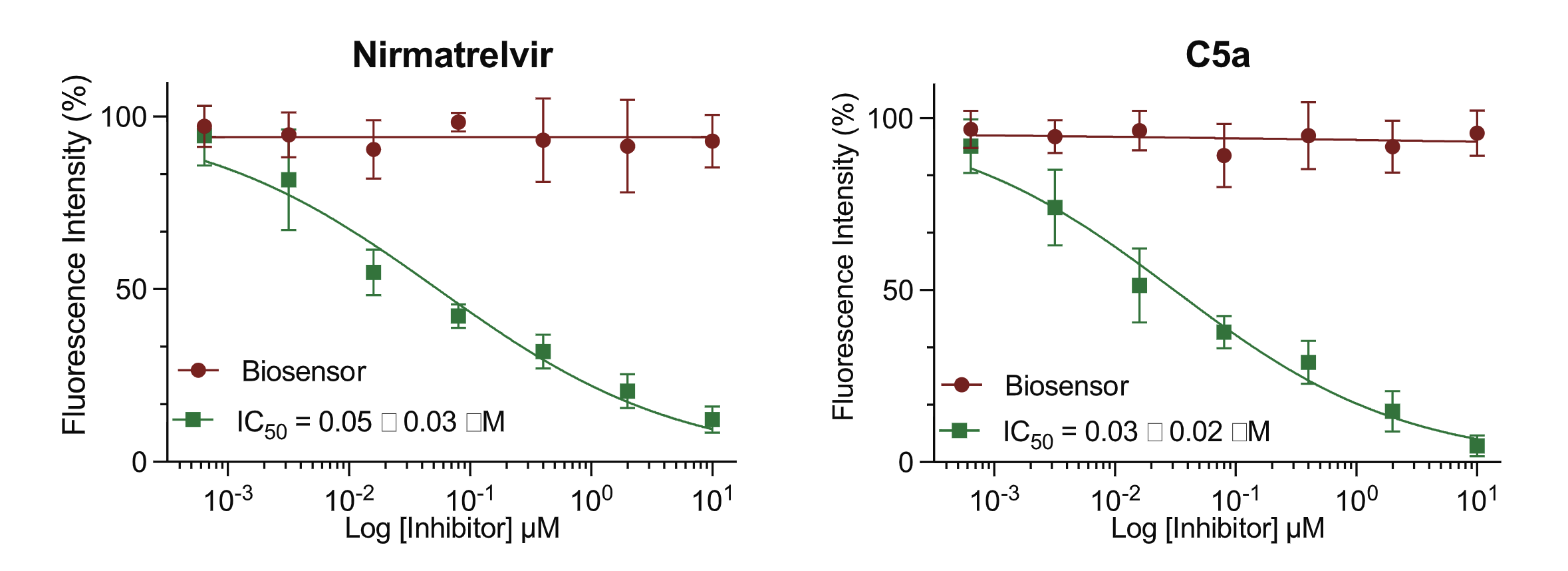
An interdisciplinary group of scientists set out to discover new 3CLpro inhibitors for emerging viruses that are resistant to Paxlovid (Pérez-Vargas et al., 2023). They used a computational approach to target the active site of the protease and arrived at a series of compounds that block the protease activity. These were first tested in biochemical assays, but to demonstrate that these compounds would work in the context of living cells, they turned to Montana Molecular’s 3CLglow assay. This assay delivers the SARS-CoV-2 3CLpro as well as a bright fluorescent biosensor for protease activity. The 3CLglow assay demonstrated that their new compound, C5a, was quite effective at blocking the protease activity, and that the compound crosses the cell membrane and effectively engages the target protease (Figure 2O; Pérez-Vargas et al., 2023)
The final proof of efficacy was done by inhibiting the replication of SARS-CoV-2 virus strains, including BA.5, BQ.1.1, and XBB.1.5. Remarkably, the EC50 values for inhibition (~20-80 nM) indicate that C5a is quite potent. C5a, or derivatives to come, could be promising alternatives to Paxlovid and could become the next generation of drugs we will inevitably need to combat the new coronavirus variants on our horizon.
“I believe we are close to seeing SARS-CoV-2 mutate enough that it will evade all remaining treatments. Our next-generation antivirals in development, such as C5/C5a, have the potential to fill significant treatment gaps in the short term associated with BA.2 variants such as BA.2.86 and to ensure future, long-term preparedness.” – CoVaRR-Net
3CLglow: A live cell assay for SARS-CoV-2 3CLpro enzyme activity and inhibition.
The 3CLglow Assay kit includes 2 BacMam viruses. One expresses the 3CLpro enzyme, the other expresses a green fluorescent biosensor for 3CLpro enzyme activity as well as a red fluorescent protein to control for compound toxicity.
- Identify protease inhibitors that target SARS-CoV-2 replication
- Cost effective screening with minimal safety requirements (BSL-1)
References
Iketani, S., Mohri, H., Culbertson, B., Hong, S. J., Duan, Y., Luck, M. I., Annavajhala, M. K., Guo, Y., Sheng, Z., Uhlemann, A.-C., Goff, S. P., Sabo, Y., Yang, H., Chavez, A., & Ho, D. D. (2023). Multiple pathways for SARS-CoV-2 resistance to nirmatrelvir. Nature, 613(7944), 558–564. https://doi.org/10.1038/s41586-022-05514-2
Joyce, R. P., Hu, V. W., & Wang, J. (2022). The history, mechanism, and perspectives of nirmatrelvir (PF-07321332): an orally bioavailable main protease inhibitor used in combination with ritonavir to reduce COVID-19-related hospitalizations. Medicinal Chemistry Research: An International Journal for Rapid Communications on Design and Mechanisms of Action of Biologically Active Agents, 31(10), 1637–1646. https://doi.org/10.1007/s00044-022-02951-6
Kiso, M., Furusawa, Y., Uraki, R., Imai, M., Yamayoshi, S., & Kawaoka, Y. (2023). In vitro and in vivo characterization of SARS-CoV-2 strains resistant to nirmatrelvir. Nature Communications, 14(1), 3952. https://doi.org/10.1038/s41467-023-39704-x
Moghadasi, S. A., Heilmann, E., Khalil, A. M., Nnabuife, C., Kearns, F. L., Ye, C., Moraes, S. N., Costacurta, F., Esler, M. A., Aihara, H., von Laer, D., Martinez-Sobrido, L., Palzkill, T., Amaro, R. E., & Harris, R. S. (2023). Transmissible SARS-CoV-2 variants with resistance to clinical protease inhibitors. Science Advances, 9(13), eade8778. https://doi.org/10.1126/sciadv.ade8778
Owen, D. R., Allerton, C. M. N., Anderson, A. S., Aschenbrenner, L., Avery, M., Berritt, S., Boras, B., Cardin, R. D., Carlo, A., Coffman, K. J., Dantonio, A., Di, L., Eng, H., Ferre, R., Gajiwala, K. S., Gibson, S. A., Greasley, S. E., Hurst, B. L., Kadar, E. P., … Zhu, Y. (2021). An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science, 374(6575), 1586–1593. https://doi.org/10.1126/science.abl4784
Pérez-Vargas, J., Worrall, L. J., Olmstead, A. D., Ton, A.-T., Lee, J., Villanueva, I., Thompson, C. A. H., Dudek, S., Ennis, S., Smith, J. R., Shapira, T., De Guzman, J., Gang, S., Ban, F., Vuckovic, M., Bielecki, M., Kovacic, S., Kenward, C., Hong, C. Y., … Jean, F. (2023). A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants. Emerging Microbes & Infections, 12(2), 2246594. https://doi.org/10.1080/22221751.2023.2246594
Smith, E., Davis-Gardner, M. E., Garcia-Ordonez, R. D., Nguyen, T.-T., Hull, M., Chen, E., Yu, X., Bannister, T. D., Baillargeon, P., Scampavia, L., Griffin, P., Farzan, M., & Spicer, T. P. (2023). High throughput screening for drugs that inhibit 3C-like protease in SARS-CoV-2. SLAS Discovery, 28(3), 95–101. https://doi.org/10.1016/j.slasd.2023.01.001
CoVaRR-Net, A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants, https://covarrnet.ca/a-novel-class-of-broad-spectrum-active-site-directed-3c-like-protease-inhibitors-with-nanomolar-antiviral-activity-against-highly-immune-evasive-sars-cov-2-omicron-subvariants/